Overview
- Metadata and data can be submitted to our platform using Microsoft Excel WorkBooks that describe related items in separate sheets.
- This section provides detailed information on how to use the WorkBooks.
- You can check out the example WorkBook we prepared for the data from Rao et. al. 2014 to familiarize yourself with the general structure.
- Based on the type of experiment(s) for which you plan to submit data, the data wranglers can provide you with an Excel WorkBook containing several WorkSheets.
- Each sheet corresponds to an Item type in our metadata database.
- The workbook provided should contain all the sheets that you may need for your submission.
- You can refer to this table for information on all the Item types available in the database.
- Each sheet should also contain all the data fields that can be submitted for that Item type.
- Depending on if you have submitted data before or if you are using shared reagents that have been submitted by other labs, you may not need to provide information on every sheet or in every field.
Organization of the Workbook
- Generally, it makes sense to begin with the left most sheet in the workbook as the sheets in a workbook are ordered so that Items that have fields that take a reference to another Item as their value appear ‘after’ i.e. to the right of that Item’s sheet in the workbook.
- A sheet for an Item starts with a row of field names.
- Absolutely required fields are marked with a leading asterisk (eg. *experiment_type). - failure to supply a value in these fields will cause an error
- The second row of the sheet indicates the type of the information expected for the fields.
- The third row includes a description of each of the fields.
- In some cases the values that you can submit for a particular field are constrained to a specific set of terms and when this is the case the possible values are shown in the fourth row.
- Any row that starts with “#” in the first column will be ignored, so you can add non-data rows for your own use.
- However, PLEASE NOTE THAT THE FIRST 2 ROWS OF A SHEET SHOULD NOT BE MODIFIED.
Excel Headers
- Field name
- Field type (string, number, array, embedded object)
- Description
- Additional info (comments and choices for fields with controlled vocabulary)
- You may notice that in some sheets there are additional commented rows that contain data values.
- These are rows corresponding to items that already exist in the database and can provide you with identifiers that you can reuse in your submission (see Referencing existing items).
- Only those items that either are associated with your lab or are already released to the public will appear in these commented data rows.
- Your data entry should begin at the first non-commented row.
Preparing Excel Workbooks
- A field can be one of a few different types;
- string
- number/integer
- array/list
- Item
- The type will be indicated in the second row.
- Most field values are strings:
- a term from a controlled vocabulary, i.e. from a constrained list of choices
- a string that identifies an Item
- a text description.
- If the field type is an array, you may enter multiple values separated by commas.
Basic field formats

Basic fields example

- There are some fields values that require specific formatting. These cases and how to identify them are described below.
Required string formatting
In some cases a field value must be formatted in a certain way or the Item will fail validation. In most cases tips on formatting requirements will be included in the Additional Info row of the spreadsheet.
Examples of these are
- Date fields - YYYY-MM-DD format.
- URLs -checked for proper URI syntax.
- patterns - checked against simple regular expressions (eg. a DNA sequence can only contain A, T, G, C or N).
- Database Cross Reference (DBxref) fields that contain identifiers that refer to external databases
- In many cases the values of these fields need to be in database_name:ID format. eg. an SRA experiment identifier ‘SRA:SRX1234567’ (see also Basic fields example above).
- In a few cases where the field takes only identifiers for one specific databases the ID alone can be entered - eg. 'targeted_genes’ field of the Target Item enter gene symbols eg. PARK2, DLG1.
Linked item fields
- Some fields in a Sheet for an Item may contain references to another Item.
- The referenced Item may be of the same or different type.
- Examples of this type of field include the ‘biosource’ field in Biosample or the ‘files’ field in the ExperimentHiC.
- The 'files' field is also an example of a list field that can take multiple values.
- You can reference an item in the excel workbooks using one of four possible ways:
- lab-specific alias
- accession
- item-type-specific identifier
- UUID
More information about these four identifiers is provided in Using aliases.
Field(s) with subobjects
Some Items can contain embedded sub-objects that are stored under a single Item field name but that contain multiple sub-fields that remain grouped together.
These are indicated in the Item spreadsheet using a ‘.’ (dot) notation.
For example the "experiment_relations" field has 2 sub-fields called "relationship_type", and "experiment". In the spreadsheet field names you will see experiment_relations.relationship_type and experiment_relations.experiment.
If the Item field is designed to store a list of embedded sub-objects, you can enter multiple sub-objects by manually creating new columns and appending incremented integers to the fields names for each new sub-object.
For example, to submit a total of three related experiments to an ExperimentHiC Item you would find the experiment_relations.relationship_type and experiment_relations.experiment columns, copy them and have total of 6 columns named:
experiment_relations.relationship_type experiment_relations.experiment experiment_relations.relationship_type-1 experiment_relations.experiment-1 experiment_relations.relationship_type-2 experiment_relations.experiment-2
and enter a valid relationship_type term and experiment identifier to each of the three pairs of columns.
Multiple linked columns for lists of embedded objects

Referencing existing items
- Ways that you can reference items that already exist in the 4DN database in your spreadsheet submission is described here.
- In some cases information for existing items will be present in the Excel Work Sheets provided for your submission.
- You can also check the existing items from collection pages that list all of them.
- The links for item lists can be constructed by
(e.g. https://data.4dnucleome.org/biosamples/ ) and the identifiers that can be used for collections are referenced in this table.
Supplementary metadata files
To submit supplementary metadata files, such as pdfs or images, use the Image or Document schemas, and include the path of the files in the *attachment* column.The path should be the full path to the supplementary file.
Experimental Replicate information
- All experiments must be part of a replicate set - even if it is a set containing only a single experiment.
- When preparing your submission you should determine how many replicate sets you will be submitting and create an entry - with an alias and preferably an informative description - for each set in the ExperimentSetReplicate sheet.
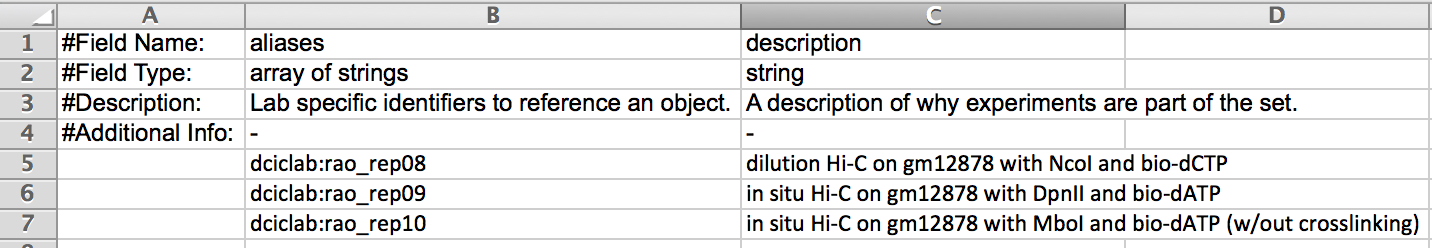
- Then when entering information about individual experiments on the specific
Experimentsheet you should:- enter the alias for the replicate set to which the experiment belongs
- indicate the bioreplicate and technical replicate number for that experiment.
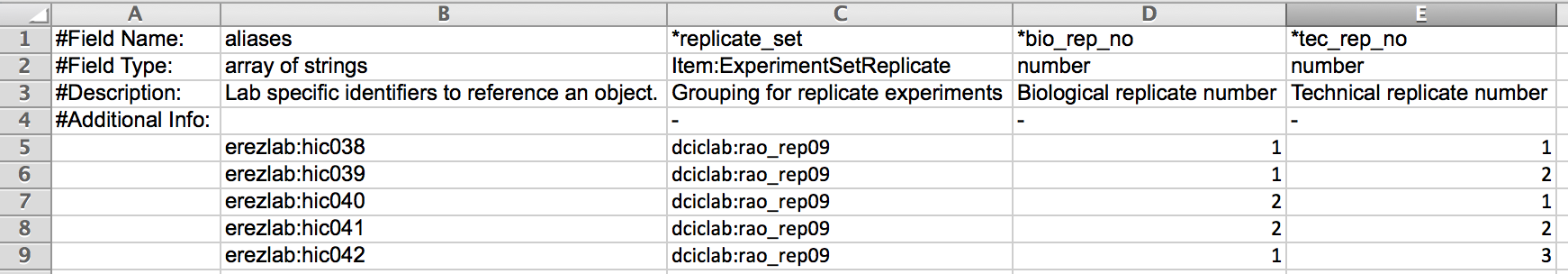
- In the example below the replicate set consists of five experiments categorized into one of two bioreplicates - bio_rep_no 1 and bio_rep_no 2, each of which contains three and two technical replicates, respectively.
Submitting Excel Workbooks
- The 4DN DCIC website has an REST API for fetching and submitting data.
- In our Submit4DN package the
import_datascript utilizes an organized bundle of REST API commands that parse the Excel workbook and submit the metadata to the database for you. - The
get_field_infoscript that is also part of the package can be used to generate the Excel workbook templates used for submission for all or a selected set of worksheets. - The package can be installed from pypi.
Installing the Submit4DN software
The Submit4DN package is registered with Pypi so installation is as simple as::
pip3 install submit4dn
If it is already installed upgrade to the latest version::
pip3 install submit4dn --upgrade
Submit4DN Source code
The source code for the submission scripts is available on github.
Note if you are attempting to run the scripts in the wranglertools directory without installing the package, then in order to get the correct sys.path you need to run the scripts from the parent directory as modules using the -m flag.:
python3 -m wranglertools.import_data filename.xls
Using import_data script for submission
- You can use
import_dataeither to upload new items or to modify metadata fields of existing items. - This script will accept the excel workbook you prepared, and will upload every new item in the sheets.
- This script is also used to upload data files to the 4DN data store - this is done in a separate step after your File metadata has been successfully uploaded.
Get access keys
You will need to generate access keys to submit data. How to get these is described here.
Workbook Submission
Testing your metadata
Before actually updating the 4DN database you can check your spreadsheet for formatting and missing required data by doing a 'dry run'.
When you run the import_data script on your metadata excel workbook without the
--updateor--patchallarguments the system will test your data for compatibility with our metadata structure and report back to you any problems.The metadata will not be submitted to the database, so you can take advantage of this feature to test your excel workbook.
import_data My_metadata.xls
Uploading (posting) & Modifying (patching) Metadata
When you submit your metadata, if a row in any sheet corresponds to a new item that has not previously been submitted to the 4DN database you will be POSTing that data via the REST API.
Most of your entries in the first submission will be POSTs. To activate posting you need to include the
--updateargument toimport_data.import_data My_metadata.xls --update
If you need to modify an existing item, you can use the patch function.
To be able to match your item to the one on the server, a pre-existing identifier must be used in the spreadsheet.
If you included an alias when you posted the item, you can use this alias to reference the existing item in the database -- uuids, @ids, or accessions can also be used to reference existing items in the database.
If you don't use the
--patchallargument when you runimport_dataand an existing entry is encountered, the script will prompt you ‘Do you wish to PATCH this item?’. You will be prompted for every existing item that is found in your workbook.The
--patchallargument will allow automatic patching of each existing item, bypassing the prompts.import_data My_metadata.xls --patchall
When your upload is aborted
If for some reason the script fails in the middle of the upload process or errors are encountered for certain items, some items will have been posted while others will have not.
When you fix the problem that caused the process to terminate, you can rerun the script using both the
--patchalland--updatearguments.Those items that had already been posted will be ‘patched’ using the data in the sheet and the items that had not been posted yet will be loaded.
import_data My_metadata.xls --patchall --update
Functionality that will allow the deletion of all the data in a single field of an existing Item exists - however this can be a potentially dangerous operation. If you determine that you need this functionality please contact us at the DCIC for more information.
Uploading files with import_data
- The 4DN databased distinguishes two main categories of files:
- files that support the metadata, such as Documents or Images
- data files for which metadata is gathered and are specified in specific File items/sheets (eg. FileFastq).
- The first category can be uploaded along with the metadata by using the “attachment” fields in the excel workbook (eg. pdf, png, doc, …) as described previously.
- The second category includes the data files that are the results of experiments, eg. fastq files from HiC experiments.
- These data files are bound to a File item with a specific type eg, FileFastq that contains relevant metadata about the specific result file.
- Metadata for a file should be submitted as part of your experiment metadata submission as described above.
- The actual file upload to the 4DN file store in the cloud will happen in a subsequent submission step. NOTE that the filename is not part of the initial File metadata submission.
- This second step will be triggered by a successful metadata submission that passes review by the 4DN DCIC.
Data File upload
To upload your files:
- use the file submission excel sheet provided
- copy paste all your file (FileFastq) aliases from your metadata excel sheet to the aliases field of the file submission sheet
- Under filename enter the full paths to your files
- use import_data with the
--patchallargument to start upload.
The DCIC automatically checks file md5sums to confirm successful upload and to ensure that there are no duplicate files in the database.
Tip
Upload using ftp is also supported, however the process currently transfers the files to your hard drive, uploads them to our system, and then deletes the copy from your local hard drive. The files are processed sequentially so you need to have at least the amount of free space on your hard drive as the size of the largest file you wish to upload. In addition, you must include your ftp login credentials in the ftp url, which is definitely not a security best practice. For these reasons, if at all possible, it is recommended to install the Submit4DN package onto the server hosting the files to be submitted and use import_data as described above. However, if that is not an option then your ftp urls should be formatted as follows:
ftp://username:password@hostname/path/to/filename
To replace a file that has already been uploaded to 4DN - that is to associate a different file with existing metadata,
- in the filename field include the new path for the existing alias
- NOTE that every time you patch with a filename (even if it is the same filename) the file will be uploaded. Please use care when including a filename in your File metadata to avoid unnecessary uploads.
- We plan to avoid this issue in future releases by pre-checking md5sums.
Generate a new Template Workbook
To create the data submission xls forms, you can use get_field_info, which is part of the Submit4DN package.
The scripts accepts the following parameters:
--keyfile access key file path (default is “home_folder/keypairs.json”) --key name of the key (default is “default”) --type use for each sheet that you want to add to the excel workbook --descriptions adds the descriptions in the second line (by default True) --enums adds the enum options in the third line (by default True) --comments adds the comments together with enums (by default False) --writexls creates the xls file (by default True) --outfile change the default file name "fields.xls" to a specified one
Examples generating a single sheet:
get_field_info --type Biosample get_field_info --type Biosample --comments get_field_info --type Biosample --comments --outfile biosample.xls
To get the complete list of relevant sheets in one workbook:
get_field_info --type all --comments --outfile AllItems.xls